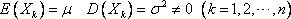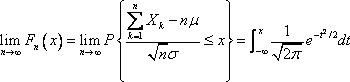1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
| class MDN:
''' Mixture Density Network which handles multi-output, full (symmetric) covariance.
Parameters
----------
n_mix : int, optional (default=5)
Number of mixtures used in the gaussian mixture model.
hidden : list, optional (default=[100, 100, 100, 100, 100])
Number of layers and hidden units per layer in the neural network.
lr : float, optional (default=1e-3)
Learning rate for the model.
l2 : float, optional (default=1e-3)
L2 regularization scale for the model weights.
n_iter : int, optional (default=1e4)
Number of iterations to train the model for
batch : int, optional (default=128)
Size of the minibatches for stochastic optimization.
imputations : int, optional (default=5)
Number of samples used in multiple imputation when handling NaN
target values during training. More samples results in a higher
accuracy for the likelihood estimate, but takes longer and may
result in overfitting. Assumption is that any missing data is
MAR / MCAR, in order to allow a multiple imputation approach.
epsilon : float, optional (default=1e-3)
Normalization constant added to diagonal of the covariance matrix.
activation : str, optional (default=relu)
Activation function applied to hidden layers.
scalerx : transformer, optional (default=IdentityTransformer)
Transformer which has fit, transform, and inverse_transform methods
(i.e. follows the format of sklearn transformers). Scales the x
values prior to training / prediction. Stored along with the saved
model in order to have consistent inputs to the model.
scalery : transformer, optional (default=IdentityTransformer)
Transformer which has fit, transform, and inverse_transform methods
(i.e. follows the format of sklearn transformers). Scales the y
values prior to training, and the output values after prediction.
Stored along with the saved model in order to have consistent
outputs from the model.
model_path : pathlib.Path, optional (default=./Weights)
Folder location to store saved models.
model_name : str, optional (default=MDN)
Name to assign to the model.
no_load : bool, optional (default=False)
If true, train a new model rather than loading a previously
trained one.
no_save : bool, optional (default=False)
If true, do not save the model when training is completed.
seed : int, optional (default=None)
Random seed. If set, ensure consistent output.
verbose : bool, optional (default=False)
If true, print various information while loading / training.
debug : bool, optional (default=False)
If true, use control flow dependencies to determine where NaN
values are entering the model. Model runs slower with this
parameter set to true.
'''
distribution = 'MultivariateNormalTriL'
def __init__(self, n_mix=5, hidden=[100]*5, lr=1e-3, l2=1e-3, n_iter=1e4,
batch=128, imputations=5, epsilon=1e-3,
activation='relu',
scalerx=None, scalery=None,
model_path='Weights', model_name='MDN',
no_load=False, no_save=False,
seed=None, verbose=False, debug=False, **kwargs):
config = initialize_random_states(seed)
config.update({
'n_mix' : n_mix,
'hidden' : list(np.atleast_1d(hidden)),
'lr' : lr,
'l2' : l2,
'n_iter' : n_iter,
'batch' : batch,
'imputations' : imputations,
'epsilon' : epsilon,
'activation' : activation,
'scalerx' : scalerx if scalerx is not None else IdentityTransformer(),
'scalery' : scalery if scalery is not None else IdentityTransformer(),
'model_path' : Path(model_path),
'model_name' : model_name,
'no_load' : no_load,
'no_save' : no_save,
'seed' : seed,
'verbose' : verbose,
'debug' : debug,
})
self.set_config(config)
for k in kwargs:
warnings.warn(f'Unused keyword given to MDN: "{k}"', UserWarning)
def _predict_chunk(self, X, return_coefs=False, use_gpu=False, **kwargs):
''' Generates estimates for the given set. X may be only a subset of the full
data, which speeds up the prediction process and limits memory consumption.
use_gpu : bool, optional (default=False)
Use the GPU to generate estimates if True, otherwise use the CPU.
'''
with tf.device('/gpu:0' if use_gpu else '/cpu:0'):
model_out = self.model( self.scalerx.transform(ensure_format(X)) )
coefs_out = self.get_coefs(model_out)
outputs = self.extract_predictions(coefs_out, **kwargs)
if return_coefs:
return outputs, [c.numpy() for c in coefs_out]
return outputs
@ignore_warnings
def predict(self, X, chunk_size=1e5, return_coefs=False, **kwargs):
'''
Top level interface to get predictions for a given dataset, which wraps _predict_chunk
to generate estimates in smaller chunks. See the docstring of extract_predictions() for
a description of other keyword parameters that can be given.
chunk_size : int, optional (default=1e5)
Controls the size of chunks which are estimated by the model. If None is passed,
chunking is not used and the model is given all of the X dataset at once.
return_coefs : bool, optional (default=False)
If True, return the estimated coefficients (prior, mu, sigma) along with the
other requested outputs. Note that rescaling the coefficients using scalerx/y
is left up to the user, as calculations involving sigma must be performed in
the basis learned by the model.
'''
chunk_size = int(chunk_size or len(X))
partial_coefs = []
partial_estim = []
for i in trange(0, len(X), chunk_size, disable=not self.verbose):
chunk_est, chunk_coef = self._predict_chunk(X[i:i+chunk_size], return_coefs=True, **kwargs)
partial_coefs.append(chunk_coef)
partial_estim.append( np.array(chunk_est, ndmin=3) )
coefs = [np.vstack(c) for c in zip(*partial_coefs)]
preds = np.hstack(partial_estim)
if return_coefs:
return preds, coefs
return preds
def extract_predictions(self, coefs, confidence_interval=None, threshold=None, avg_est=False):
'''
Function used to extract model predictions from the given set of
coefficients. Users should call the predict() method instead, if
predictions from input data are needed.
confidence_interval : float, optional (default=None)
If a confidence interval value is given, then this function
returns (along with the predictions) the upper and lower
{confidence_interval*100}% confidence bounds around the prediction.
threshold : float, optional (default=None)
If set, the model outputs the maximum prior estimate when the prior
probability is above this threshold; and outputs the average estimate
when below the threshold. Any passed value should be in the range (0, 1],
though the sign of the threshold can be negative in order to switch the
estimates (i.e. negative threshold would output average estimate when prior
is greater than the (absolute) value).
avg_est : bool, optional (default=False)
If true, model outputs the prior probability weighted mean as the
estimate. Otherwise, model outputs the maximum prior estimate.
'''
assert(confidence_interval is None or (0 < confidence_interval < 1)), 'confidence_interval must be in the range (0,1)'
assert(threshold is None or (0 < threshold <= 1)), 'threshold must be in the range (0,1]'
target = ('avg' if avg_est else 'top') if threshold is None else 'threshold'
output = getattr(self, f'_get_{target}_estimate')(coefs)
scale = lambda x: self.scalery.inverse_transform(x.numpy())
if confidence_interval is not None:
assert(threshold is None), f'Cannot calculate confidence on thresholded estimates'
confidence = getattr(self, f'_get_{target}_confidence')(coefs, confidence_interval)
upper_bar = output + confidence
lower_bar = output - confidence
return scale(output), scale(upper_bar), scale(lower_bar)
return scale(output)
@ignore_warnings
def fit(self, X, Y, output_slices=None, **kwargs):
with get_device(self.config):
checkpoint = self.model_path.joinpath('checkpoint')
if checkpoint.exists() and not self.no_load:
if self.verbose: print(f'Restoring model weights from {checkpoint}')
self.load()
elif self.no_load and X is None:
raise Exception('Model exists, but no_load is set and no training data was given.')
elif X is not None and Y is not None:
self.scalerx.fit( ensure_format(X), ensure_format(Y) )
self.scalery.fit( ensure_format(Y) )
datasets = kwargs['datasets'] = kwargs.get('datasets', {})
datasets.update({'train': {'x' : X, 'y': Y}})
for key, data in datasets.items():
if data['x'] is not None:
datasets[key].update({
'x_t' : self.scalerx.transform( ensure_format(data['x']) ),
'y_t' : self.scalery.transform( ensure_format(data['y']) ),
})
assert(np.isfinite(datasets['train']['x_t']).all()), 'NaN values found in X training data'
self.update_config({
'output_slices' : output_slices or {'': slice(None)},
'n_inputs' : datasets['train']['x_t'].shape[1],
'n_targets' : datasets['train']['y_t'].shape[1],
})
self.build()
callbacks = []
model_kws = {
'batch_size' : self.batch,
'epochs' : max(1, int(self.n_iter / max(1, len(X) / self.batch))),
'verbose' : 0,
'callbacks' : callbacks,
}
if self.verbose:
callbacks.append( TqdmCallback(model_kws['epochs'], data_size=len(X), batch_size=self.batch) )
if self.debug:
callbacks.append( tf.keras.callbacks.TensorBoard(histogram_freq=1, profile_batch=(2,60)) )
if 'args' in kwargs:
if getattr(kwargs['args'], 'plot_loss', False):
callbacks.append( PlottingCallback(kwargs['args'], datasets, self) )
if getattr(kwargs['args'], 'save_stats', False):
callbacks.append( StatsCallback(kwargs['args'], datasets, self) )
if getattr(kwargs['args'], 'best_epoch', False):
if 'valid' in datasets and 'x_t' in datasets['valid']:
model_kws['validation_data'] = (datasets['valid']['x_t'], datasets['valid']['y_t'])
callbacks.append( ModelCheckpoint(self.model_path) )
self.model.fit(datasets['train']['x_t'], datasets['train']['y_t'], **model_kws)
if not self.no_save:
self.save()
else:
raise Exception(f"No trained model exists at: \n{self.model_path}")
return self
def build(self):
layer_kwargs = {
'activation' : self.activation,
'kernel_regularizer' : tf.keras.regularizers.l2(self.l2),
'bias_regularizer' : tf.keras.regularizers.l2(self.l2),
}
mixture_kwargs = {
'n_mix' : self.n_mix,
'n_targets' : self.n_targets,
'epsilon' : self.epsilon,
}
mixture_kwargs.update(layer_kwargs)
create_layer = lambda inp, out: tf.keras.layers.Dense(out, input_shape=(inp,), **layer_kwargs)
model_layers = [create_layer(inp, out) for inp, out in zip([self.n_inputs] + self.hidden[:-1], self.hidden)]
output_layer = MixtureLayer(**mixture_kwargs)
optimizer = tf.keras.optimizers.Adam(self.lr)
self.model = tf.keras.Sequential(model_layers + [output_layer], name=self.model_name)
self.model.compile(loss=self.loss, optimizer=optimizer, metrics=[])
@tf.function
def loss(self, y, output):
prior, mu, scale = self._parse_outputs(output)
dist = getattr(tfp.distributions, self.distribution)(mu, scale)
prob = tfp.distributions.Categorical(probs=prior)
mix = tfp.distributions.MixtureSameFamily(prob, dist)
def impute(mix, y, N):
return tf.reduce_mean([
mix.log_prob( tf.where(tf.math.is_nan(y), mix.sample(), y) )
for _ in range(N)], 0)
likelihood = mix.log_prob(y)
return tf.reduce_mean(-likelihood) + tf.add_n([0.] + self.model.losses)
def __call__(self, inputs):
return self.model(inputs)
def get_config(self):
return self.config
def set_config(self, config, *args, **kwargs):
self.config = {}
self.update_config(config, *args, **kwargs)
def update_config(self, config, keys=None):
if keys is not None:
config = {k:v for k,v in config.items() if k in keys or k not in self.config}
self.config.update(config)
for k, v in config.items():
setattr(self, k, v)
def save(self):
self.model_path.mkdir(parents=True, exist_ok=True)
store_pkl(self.model_path.joinpath('config.pkl'), self.get_config())
self.model.save_weights(self.model_path.joinpath('checkpoint'))
def load(self):
self.update_config(read_pkl(self.model_path.joinpath('config.pkl')), ['scalerx', 'scalery', 'tf_random', 'np_random'])
tf.random.set_global_generator(self.tf_random)
if not hasattr(self, 'model'): self.build()
self.model.load_weights(self.model_path.joinpath('checkpoint')).expect_partial()
def get_coefs(self, output):
prior, mu, scale = self._parse_outputs(output)
return prior, mu, self._covariance(scale)
def _parse_outputs(self, output):
prior, mu, scale = tf.split(output, [self.n_mix, self.n_mix * self.n_targets, -1], axis=1)
prior = tf.reshape(prior, shape=[-1, self.n_mix])
mu = tf.reshape(mu, shape=[-1, self.n_mix, self.n_targets])
scale = tf.reshape(scale, shape=[-1, self.n_mix, self.n_targets, self.n_targets])
return prior, mu, scale
def _covariance(self, scale):
return tf.einsum('abij,abjk->abik', tf.transpose(scale, perm=[0,1,3,2]), scale)
'''
Estimate Generation
'''
def _calculate_top(self, prior, values):
vals, idxs = tf.nn.top_k(prior, k=1)
idxs = tf.stack([tf.range(tf.shape(idxs)[0]), tf.reshape(idxs, [-1])], axis=-1)
return tf.gather_nd(values, idxs)
def _get_top_estimate(self, coefs, **kwargs):
prior, mu, _ = coefs
return self._calculate_top(prior, mu)
def _get_avg_estimate(self, coefs, **kwargs):
prior, mu, _ = coefs
return tf.reduce_sum(mu * tf.expand_dims(prior, -1), 1)
def _get_threshold_estimate(self, coefs, threshold=0.5):
top_estimate = self.get_top_estimate(coefs)
avg_estimate = self.get_avg_estimate(coefs)
prior, _, _ = coefs
return tf.compat.v2.where(tf.expand_dims(tf.math.greater(tf.reduce_max(prior, 1) / threshold, tf.math.sign(threshold)), -1), top_estimate, avg_estimate)
'''
Confidence Estimation
'''
def _calculate_confidence(self, sigma, level=0.9):
s, u, v = tf.linalg.svd(sigma)
rho = 2**0.5 * tf.math.erfinv(level ** (1./self.n_targets))
return tf.cast(rho, tf.float32) * 2 * s ** 0.5
def _get_top_confidence(self, coefs, level=0.9):
prior, mu, sigma = coefs
top_sigma = self._calculate_top(prior, sigma)
return self._calculate_confidence(top_sigma, level)
def _get_avg_confidence(self, coefs, level=0.9):
prior, mu, sigma = coefs
avg_estim = self.get_avg_estimate(coefs)
avg_sigma = tf.reduce_sum(tf.expand_dims(tf.expand_dims(prior, -1), -1) *
(sigma + tf.matmul(tf.transpose(mu - tf.expand_dims(avg_estim, 1), (0,2,1)),
mu - tf.expand_dims(avg_estim, 1))), axis=1)
return self._calculate_confidence(avg_sigma, level)
class MixtureLayer(tf.keras.layers.Layer):
def __init__(self, n_mix, n_targets, epsilon, **layer_kwargs):
super(MixtureLayer, self).__init__()
layer_kwargs.pop('activation', None)
self.n_mix = n_mix
self.n_targets = n_targets
self.epsilon = tf.constant(epsilon)
self._layer = tf.keras.layers.Dense(self.n_outputs, **layer_kwargs)
@property
def layer_sizes(self):
''' Sizes of the prior, mu, and (lower triangle) scale matrix outputs '''
sizes = [1, self.n_targets, (self.n_targets * (self.n_targets + 1)) // 2]
return self.n_mix * np.array(sizes)
@property
def n_outputs(self):
''' Total output size of the layer object '''
return sum(self.layer_sizes)
def call(self, inputs):
prior, mu, scale = tf.split(self._layer(inputs), self.layer_sizes, axis=1)
prior = tf.nn.softmax(prior, axis=-1) + tf.constant(1e-9)
mu = tf.stack(tf.split(mu, self.n_mix, 1), 1)
scale = tf.stack(tf.split(scale, self.n_mix, 1), 1)
scale = tfp.math.fill_triangular(scale, upper=False)
norm = tf.linalg.diag(tf.ones((1, 1, self.n_targets)))
sigma = tf.einsum('abij,abjk->abik', tf.transpose(scale, perm=[0,1,3,2]), scale)
sigma+= self.epsilon * norm
scale = tf.linalg.cholesky(sigma)
return tf.keras.layers.concatenate([
tf.reshape(prior, shape=[-1, self.n_mix]),
tf.reshape(mu, shape=[-1, self.n_mix * self.n_targets]),
tf.reshape(scale, shape=[-1, self.n_mix * self.n_targets ** 2]),
])
|